Applied Data Science: Subgroup Discovery on Mushrooms
My last publication was on Subgroup Discovery for Sequences (you can access it freely here). However, in Data Science community, a lot of people are not aware of what “Subgroup Discovery” or “Pattern Mining” is. So let’s see on a quick pratical example how to use it : knowing if Mushrooms are poisonous.
What is Subgroup Discovery ?
Subgroup Discovery, Emerging Patterns, Contrast Set, or Discriminative Pattern Mining all refer to the same idea: finding patterns that are discriminative of a target class. In other words, the aim is to find predictive interpretable rules of a class. As an example, Herrera et al used Subgroup Discovery in the context of a pyschiatric emergency department. They found rules like:
- If Sex=Male and DAY=Monday -> Suicide
- If Sex=Female and (DAY=SUNDAY or DAY=MONDAY) and TIME=LATE_EVENING -> Suicide
Of course these rules are not correct 100% of the time, but they tell you that when a pattern appears, there are more chances that the class appears too.
This is interesting for two reasons:
- Understanding your data in a way that is interpretable by an expert.
- Using those patterns to improve classification or regression algorithms. Indeed, as they are discriminative of a target class, you can use them as features to improve classical supervised learning.
Subgroup discovery can then be used to improve your system, thanks to interpretability: knowing that people have more suicidal thoughts on Sunday and Monday, particulary at night, you can engage more psychology support workers during those periods of time in the department, for example.
Let’s try it on Mushroom
Mushroom is a famous dataset which contains characteristics of different species of mushrooms: its odor, color, habitat etc. More importantly, there is also the information if a mushroom is edible, or not.
This will be our target class: when using a subgroup discovery algorithm, we consider a dataset and a target class, and the algorithm returns a set of rules discriminative of this class. Here, we are looking for patterns discriminative of Poisonous mushrooms. In other words, we want to find the conjunction of features that are characteristics of poisonous mushrooms.
First, let’s install pysubgroup package, which is an implementation of several subgroup discovery algorithms, in Python:
pip install pysubgroup
import pysubgroup as ps
import pandas as pd
Let’s take a look at the dataset.
data = pd.read_csv("./mushroom.csv")
print(data.describe())
EDIBLE CAP-SHAPE CAP-SURFACE CAP-COLOR BRUISES ODOR GILL-ATTACHEMENT \
count 8417 8416 8416 8416 8416 8416 8416
unique 3 6 4 10 2 9 2
top EDIBLE CONVEX SCALY BROWN NO NONE FREE
freq 4488 3796 3268 2320 5040 3808 8200
GILL-SPACING GILL-SIZE GILL-COLOR ... STALK-SURFACE-BELOW-RING \
count 8416 8416 8416 ... 8416
unique 2 2 12 ... 4
top CLOSE BROAD BUFF ... SMOOTH
freq 6824 5880 1728 ... 5076
STALK-COLOR-ABOVE-RING STALK-COLOR-BELOW-RING VEIL-TYPE VEIL-COLOR \
count 8416 8416 8416 8416
unique 9 9 1 4
top WHITE WHITE PARTIAL WHITE
freq 4744 4640 8416 8216
RING-NUMBER RING-TYPE SPORE-PRINT-COLOR POPULATION HABITAT
count 8416 8416 8416 8416 8416
unique 3 5 9 6 7
top ONE PENDANT WHITE SEVERAL WOODS
freq 7768 3968 2424 4064 3160
[4 rows x 23 columns]
We now have to specify the target class: in our case, it’s the column ‘EDIBLE’, when it takes the value ‘POISONOUS’. Mind that we should remove this column from the data (the set of features), or otherwise it will be considered as a feature, resulting in rules like POISONOUS -> POISONOUS.
target = ps.BinaryTarget ('EDIBLE', 'POISONOUS')
searchspace = ps.create_selectors(data, ignore=['EDIBLE'])
Then we have to create a Subgroup Discovery Task. In particular, we have to specify three parameters:
- the number of rules we want to extract (result_set_size),
- the maximum size of the rule (depth),
- the quality measure. If you do not know what a quality measure is and which one is better for your task, you can take the Weighted Relative Accuracy (WRAcc), which is one of the most popular of the domain.
task = ps.SubgroupDiscoveryTask (data, target, searchspace,
result_set_size=5, depth=5, qf=ps.WRAccQF())
Finally, we have to choose an algorithm to mine the rules. By default we can use the popular beam search.
results = ps.BeamSearch().execute(task)
Finally, we print the rules we have got:
results.to_dataframe()
0.19389014936350082: <<T: EDIBLE=POISONOUS; D: BRUISES=NO AND GILL-SPACING=CLOSE AND VEIL-TYPE=PARTIAL AND VEIL-COLOR=WHITE>>
0.19389014936350082: <<T: EDIBLE=POISONOUS; D: BRUISES=NO AND GILL-SPACING=CLOSE AND VEIL-COLOR=WHITE>>
0.19236944009552903: <<T: EDIBLE=POISONOUS; D: BRUISES=NO AND GILL-SPACING=CLOSE AND GILL-ATTACHEMENT=FREE AND VEIL-COLOR=WHITE>>
0.19236944009552903: <<T: EDIBLE=POISONOUS; D: BRUISES=NO AND GILL-SPACING=CLOSE AND GILL-ATTACHEMENT=FREE>>
0.19236944009552903: <<T: EDIBLE=POISONOUS; D: BRUISES=NO AND GILL-SPACING=CLOSE AND VEIL-TYPE=PARTIAL AND GILL-ATTACHEMENT=FREE>>
It is important to know that the WRAcc takes its values in a range [-0.25;0.25] on a balanced dataset. Therefore, a value of 0.1938 is very good: this means that this pattern is highly discriminative of poisonous mushrooms.
Let’s take a look at the first example that we have got. We learn that if a mushroom has a close gill-spacing, a veil-type partial and white, and has no bruises, then very likely, it is a poisonous one.
This is perfectly interpretable for an expert. The following picture shows what are the gill-spacing (or Hymenium here) and the Veil.
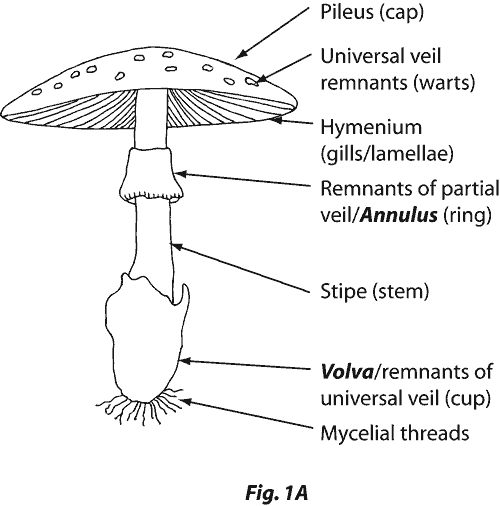
Let’s take an example with the famous Amanita phalloides. As you can see on the picture below, this mushroom has no bruises (I guess ? I am not an expert in mushrooms actually :) ), a close gill-spacing, and a white and partial veil. The rule tells you it is probably poisonous, and it is: Amanita phalloides is one of the most toxic mushrooms !
That’s it, we have extracted useful knowledge from our dataset, and we can now use it to better understand our system.
Note: The documentation of pysubgroup is lacking, but hopefully it will improve in the future.
Note: There are also other ways to extract interpretable rules, for example training a decision tree and extracting the path taken in the tree can give a pattern explaining the prediction. Clustering can also group similar elements, and finding frequent pattern between them can create interpretable rules.
The advantage of subgroup discovery over those methods is that it has been made to give those rules, whereas in those other methods it is not the main purpose of the algorithm. Here, you have more control over what kind of rules you want to propose to the end-user, particulary because you can choose the Quality Measure you want to use. In this formalism you can also use exhaustive algorithm to list all possible rules, which is not the case in a decision tree, greedy by nature.